

The toxEval R-package includes a set of functions to
analyze, visualize, and organize measured concentration data as it
relates to https://www.epa.gov/comptox-tools/toxicity-forecasting-toxcast
or other user-selected chemical-biological interaction benchmark data
such as water quality criteria. The intent of these analyses is to
develop a better understanding of the potential biological relevance of
environmental chemistry data. Results can be used to prioritize which
chemicals at which sites may be of greatest concern. These methods are
meant to be used as a screening technique to predict potential for
biological influence from chemicals that ultimately need to be validated
with direct biological assays.
The functions within this package allow great flexibly for exploring
the potential biological affects of measured chemicals. Also included in
the package is a browser-based application made from the
Shiny R-package (the app). The app is based on functions
within the R-package and includes many convenient analyses and
visualization options for users to choose. Use of the functions within
the R-package allows for additional flexibility within the functions
beyond what the app offers and provides options for the user to interact
more directly with the data. The overview in this document focuses on
the R-package.
To install the toxEval package, you must be using R 3.0 or greater and run the following command:
install.packages("toxEval")To get cutting-edge changes, install from GitHub using the
remotes packages:
library(remotes)
install_gitlab("water/toxEval",
host = "code.usgs.gov",
build_vignettes = TRUE,
build_opts = c("--no-resave-data",
"--no-manual"),
dependencies = TRUE)Installation instructions are below. To quickly get going in
toxEval, run:
library(toxEval)
#> For more information:
#> https://doi-usgs.github.io/toxEval/
#> ToxCast database: version 4.1explore_endpoints()

Then click on the “Load Example Data” in the upper right corner. This loads the example data that is found here:
file.path(system.file("extdata", package="toxEval"), "OWC_data_fromSup.xlsx")Once the data is loaded in the app, sample R code is shown below each tab. This can be copied into the R console (once the app is stopped…) to use as a base for exploring the package directly in R.
Alternatively, an example workflow is shown here (also using example data provided in the package):
library(toxEval)
path_to_file <- file.path(system.file("extdata", package="toxEval"), "OWC_data_fromSup.xlsx")
tox_list <- create_toxEval(path_to_file)
ACClong <- get_ACC(tox_list$chem_info$CAS)
ACClong <- remove_flags(ACClong)
cleaned_ep <- clean_endPoint_info(end_point_info)
filtered_ep <- filter_groups(cleaned_ep,
groupCol = 'intended_target_family',
remove_groups = c('Background Measurement','Undefined'))
chemicalSummary <- get_chemical_summary(tox_list,
ACClong,
filtered_ep)
######################################
chem_class_plot <- plot_tox_boxplots(chemicalSummary,
category = 'Chemical Class')
chem_class_plot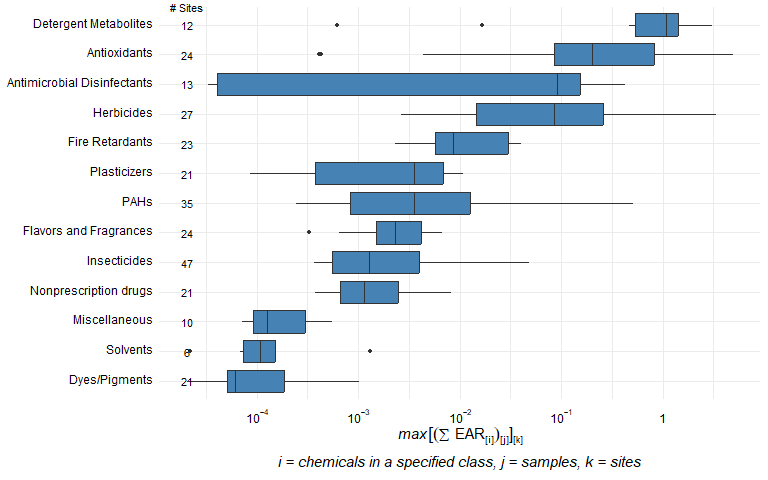
######################################
plot_stacks <- plot_tox_stacks(chemicalSummary,
tox_list$chem_site,
category = "Chemical Class")
plot_stacks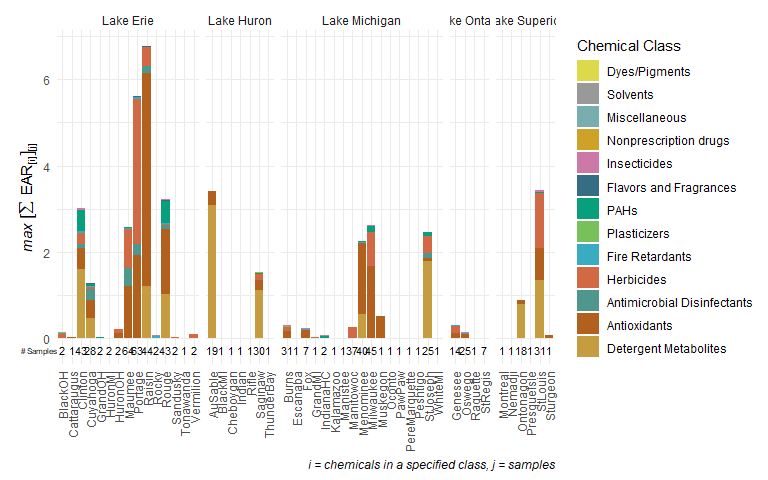
######################################
plot_heat <- plot_tox_heatmap(chemicalSummary,
tox_list$chem_site,
category = "Chemical Class",
font_size = 7)
plot_heat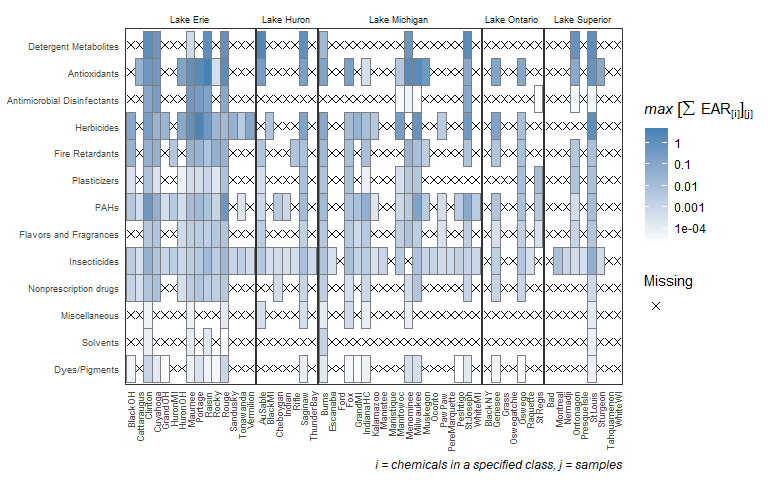
This code opens up the example file, loads it into a
toxEval object, grabs the pertinent ToxCast information,
and creates a “chemicalSummary” data frame that is used in many of the
plot and table functions.
There are 4 vignettes to help introduce and navigate the
toxEval package:
| Name | R command | Description |
|---|---|---|
| Introduction | vignette("Introduction", package="toxEval") |
Introduction to the toxEval |
| Basic Workflow | vignette("basicWorkflow", package="toxEval") |
Quickstart guide to get overview of available functions |
| Prepare Data | vignette("PrepareData", package="toxEval") |
Guide to preparing your data for toxEval analysis |
| Shiny App Guide | vignette("shinyApp", package="toxEval") |
Guide to the toxEval shiny application |
Please consider reporting bugs and asking questions on the Issues page: https://github.com/DOI-USGS/toxEval/issues
We want to encourage a warm, welcoming, and safe environment for contributing to this project. See the code of conduct for more information.
The Water and Environmental Health Mission Areas of the USGS, as well
as the Great Lakes Restoration Initiative (GLRI) has supported the
development of the toxEval R-package. Further maintenance
is expected to be stable through September 2025. Resources are available
primarily for maintenance and responding to user questions. Priorities
on the development of new features are determined by the
toxEval development team.
Funding for toxEval is secured through summer 2025,
after which bug fixes & new features will be minimal.
To run the toxEval app:
library(toxEval)
explore_endpoints()citation(package = "toxEval")
#> To cite package 'toxEval' in publications use:
#>
#> DeCicco L, Corsi S, Villeneuve D, Blackwell B, Ankley G (2024).
#> _toxEval: Exploring Biological Relevance of Environmental Chemistry
#> Observations_. R package version 1.4.0.
#>
#> A BibTeX entry for LaTeX users is
#>
#> @Manual{,
#> title = {toxEval: Exploring Biological Relevance of Environmental Chemistry
#> Observations},
#> author = {Laura DeCicco and Steven Corsi and Daniel Villeneuve and Brett Blackwell and Gerald Ankley},
#> year = {2024},
#> note = {R package version 1.4.0},
#> }